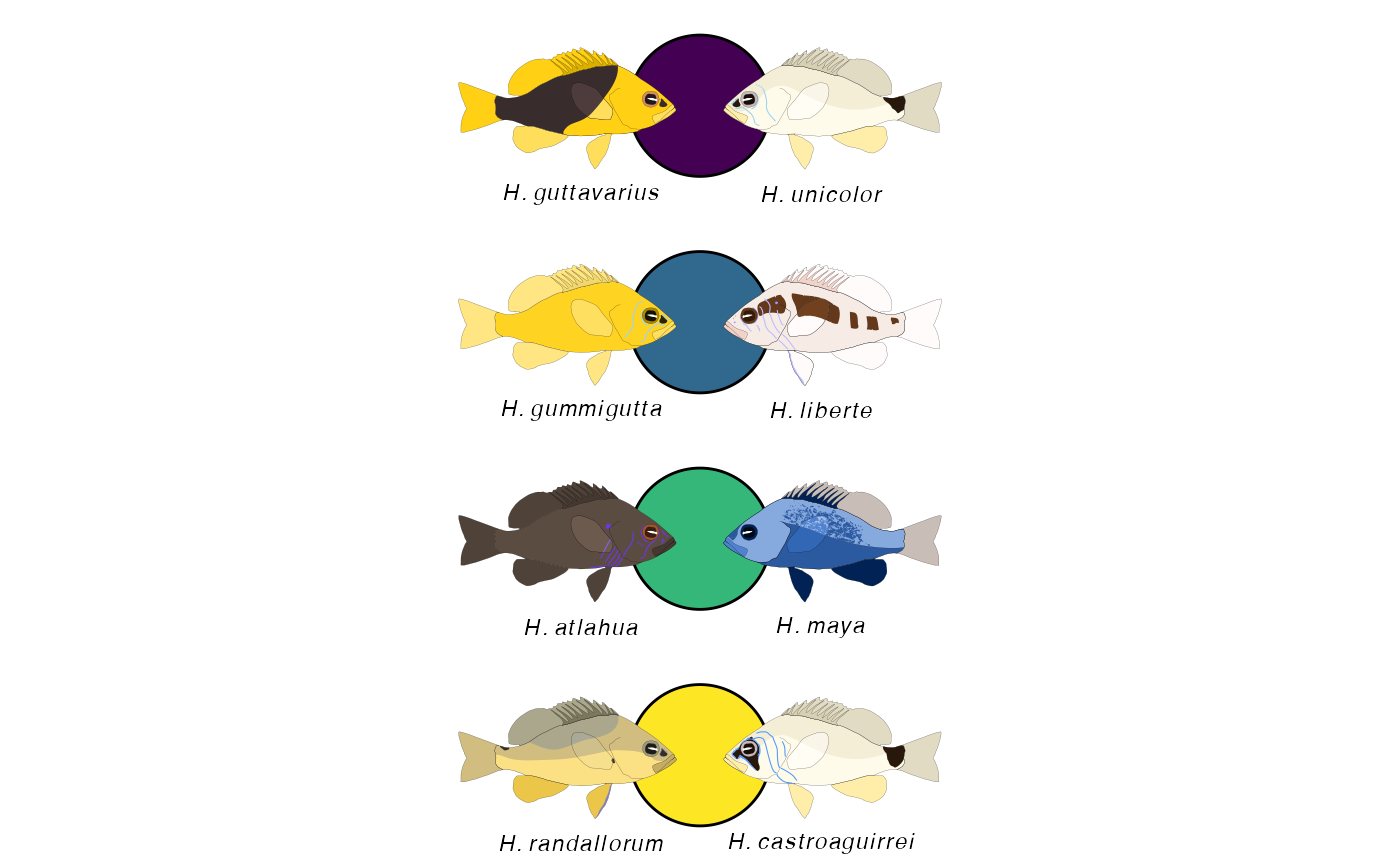
Constructs a legend of paired hamlets with two fills
Source:R/hypoimg_basics.R
hypo_legend_pair_split.Rdhypo_legend_pair_split combines several paired hamlet legend elements
hypo_legend_pair_split( left, right, color_map_left, color_map_right, circle_color = NA, circle_lwd = 0.5, plot_names = FALSE, plot_name_size = 3, font_family = "sans", ncol = 1, plot = TRUE )
Arguments
| left | string vector (manatory), can only contain available species |
|---|---|
| right | string vector (manatory), can only contain available species |
| color_map_left | color vector (a string, optional), the color map (must match the species vector in length) |
| color_map_right | color vector (a string, optional), the color map (must match the species vector in length) |
| circle_color | color scalar (a string, optional), the color of the background circle outline |
| circle_lwd | numeric scalar (optional), the width of the background circle outlines |
| plot_names | logical scalar (optional), should the species label be added? |
| plot_name_size | numeric scalar (optional), the species label size |
| font_family | string scalar (optional), the species label font family |
| ncol | interger, number of columns |
| plot | logical scalar (optional), toggle the output to be either a plot or a list of plots |
Details
The function hypo_legend_pair constructs a paired hamlet legend
from two vectors of hamlet species and a choosen color map (matching in length).
Hamlet species labels can optionally be included.
Available species: "aberrans", "atlahua", "castroaguirrei", "chlorurus", "ecosur", "floridae", "gemma", "gummigutta", "guttavarius", "indigo", "liberte", "maculiferus", "maya", "nigricans", "providencianus", "puella", "randallorum", "tan", "unicolor"
See also
Examples
clr <- viridis::viridis(4) left <- c('unicolor', 'liberte', 'maya', 'castroaguirrei') right <- c('guttavarius', 'gummigutta', 'atlahua', 'randallorum') hypo_legend_pair(left= left, right = right, color_map = clr, circle_color = 'black', plot_names = TRUE)